Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Adachi, Motoyasu; Hatanaka, Takaaki*; Ito, Yuji*; Hidaka, Koshi*; Tsuda, Yuko*; Kiso, Yoshiaki*; Kuroki, Ryota
no journal, ,
HIV protease is known as a drug target protein. It is important to clear relationship between structural data and kinetic and physicochemical parameters for drug design. We prepared single-chained enzyme linked by two amino acids and cross-liked enzyme bridged by disulfide bond. The two single-chained enzymes were expressed as inclusion body and refolded by dilution method. The purified enzyme complexed with inhibitor KNI-272 was crystallized, and solved the crystal structures. The designed sc- and cl-HIV-PR will be useful for evaluate the affinity of newly designed inhibitors from kinetic and thermodynamic point of view. Finally, we also report the results of analysis in affinity of inhibitors by surface plasmon resonance.
Kiyanagi, Ryoji; Ohara, Takashi*; Kawasaki, Takuro; Oikawa, Kenichi; Tamura, Itaru; Kaneko, Koji; Nakao, Akiko*; Hanashima, Takayasu*; Munakata, Koji*; Kimura, Hiroyuki*; et al.
no journal, ,
 reductase
reductaseYamada, Mitsugu; Tamada, Taro; Matsumoto, Fumiko; Takeda, Kazuki*; Kimura, Shigenobu*; Kuroki, Ryota; Miki, Kunio*
no journal, ,
NADH-cytochrome  reductase (b5R; EC 1.6.2.2) is a flavoprotein and catalyses two-electron transfer from NADH to cytochrome
reductase (b5R; EC 1.6.2.2) is a flavoprotein and catalyses two-electron transfer from NADH to cytochrome  through FAD. Here, we present crystal structures of fully reduced and two re-oxidized forms of b5Rs determined. The crystal structure of fully reduced b5R showed that the relative location of two domains slightly shifted in comparison with that of oxidized form. This shift allowed to create a new hydrogen bonding interaction between N5 atom of isoalloxazine ring and hydroxyl oxygen atom of Thr66. The isoalloxazine ring of FAD in fully reduced form is flat and stacked with the nicotinamide ring of bound NAD
through FAD. Here, we present crystal structures of fully reduced and two re-oxidized forms of b5Rs determined. The crystal structure of fully reduced b5R showed that the relative location of two domains slightly shifted in comparison with that of oxidized form. This shift allowed to create a new hydrogen bonding interaction between N5 atom of isoalloxazine ring and hydroxyl oxygen atom of Thr66. The isoalloxazine ring of FAD in fully reduced form is flat and stacked with the nicotinamide ring of bound NAD . Moreover, two re-oxidized forms (form-1 and 2) were prepared using fully reduced crystals by exposure to the air. The electron densities for the nicotinamide ring of NAD
. Moreover, two re-oxidized forms (form-1 and 2) were prepared using fully reduced crystals by exposure to the air. The electron densities for the nicotinamide ring of NAD was ambiguous in form-1 and was completely disappeared in form-2. These results suggested that re-oxidization follows a two-step mechanism in which nicotinamide moiety is released firstly and then whole NAD
was ambiguous in form-1 and was completely disappeared in form-2. These results suggested that re-oxidization follows a two-step mechanism in which nicotinamide moiety is released firstly and then whole NAD is released.
is released.
 -Lactamase TOHO-1 determined by combined high-resolution neutron and X-ray crystallography
-Lactamase TOHO-1 determined by combined high-resolution neutron and X-ray crystallographyKurihara, Kazuo; Sunami, Tomoko; Yamada, Mitsugu; Nitanai, Yasushi*; Okazaki, Nobuo; Adachi, Motoyasu; Tamada, Taro; Shimamura, Tatsuro*; Miyano, Masashi*; Ishii, Yoshikazu*; et al.
no journal, ,
To help resolve questions regarding the catalytic activity of  -lactamase, the crystal structure of an unliganded form of the
-lactamase, the crystal structure of an unliganded form of the  -lactamase Toho-1 with double mutation R274N/R276N (Toho-1/NN) has been determined by the use of high-resolution neutron and X-ray diffraction data. A large single crystal of Toho-1/NN with a dimension of 2.6
-lactamase Toho-1 with double mutation R274N/R276N (Toho-1/NN) has been determined by the use of high-resolution neutron and X-ray diffraction data. A large single crystal of Toho-1/NN with a dimension of 2.6  2.5
2.5  1.3 mm
1.3 mm was used to collect 100 K neutron diffraction data to 1.5
was used to collect 100 K neutron diffraction data to 1.5  resolution and X-ray diffraction data to 1.4
resolution and X-ray diffraction data to 1.4  resolution. The structural model of Toho-1/NN was refined to an R-factor of 19.7% using a program PHENIX. The structure showed that Glu166, a catalytic residue of Toho-1, was protonated even at pH 7 nonetheless for the close location to the positively charged side chain amino group (-NH3
resolution. The structural model of Toho-1/NN was refined to an R-factor of 19.7% using a program PHENIX. The structure showed that Glu166, a catalytic residue of Toho-1, was protonated even at pH 7 nonetheless for the close location to the positively charged side chain amino group (-NH3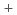 ) of Lys73. It is also found that there is a hydration water network bridging between the protonated Glu166 and the oxyanion hole comprising two main chain nitrogen atoms of Ser70 and Ser237. The neutron structure analysis also revealed the clear configuration of the proposed catalytic water molecule bridging Glu166 and Ser70. These observations are important to understand the catalytic action of
) of Lys73. It is also found that there is a hydration water network bridging between the protonated Glu166 and the oxyanion hole comprising two main chain nitrogen atoms of Ser70 and Ser237. The neutron structure analysis also revealed the clear configuration of the proposed catalytic water molecule bridging Glu166 and Ser70. These observations are important to understand the catalytic action of  -lactamase Toho-1.
-lactamase Toho-1.